Faculty Profile
 Madhan Tirumalai
Madhan Tirumalai
Research Assistant Professor
Department of Biology and Biochemistry
Office: Houston Science Center, 421
Contact: mrtirum2@central.uh.edu - 713-743-8365
- Microbial evolution and genomics
- Directed evolution (of microbes)
- Environmental microbiology
- Bacterial dormancy and mechanisms related to the same]
- Mechanisms of microbial resistance to stress, including radiation, antibiotics, and xenobiotics
- Microbiology of human-built environments
- Astrobiology & Origins of Life
- Space Microbiology – microbes of spaceflight and planetary protection concern
- Phage insertion dynamics and genome rearrangement dynamics
- Structure, function and evolution of the molecular machine ribosome, and nucleic acid RNA
- Bottom-up prebiotic chemistry – exploring RNA sequence space using dynamic combinatorial chemistry
Projects
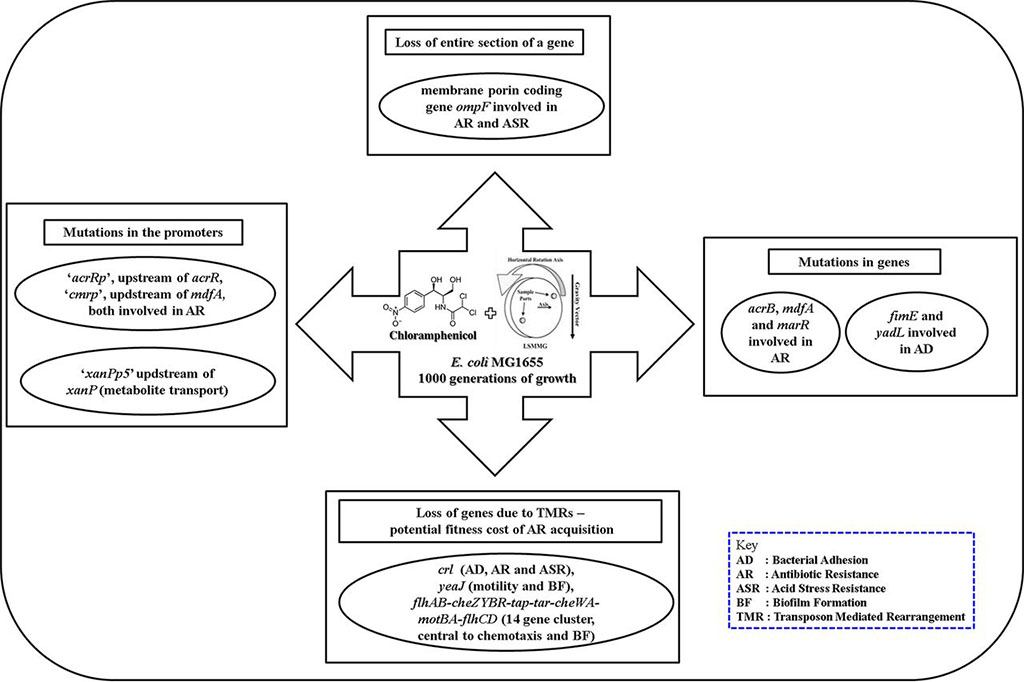
Directed evolution of bacteria under simulated microgravity and background antibiotic exposure: This has relevance to how microbes evolve and adapt to the microgravity environment found in human-made habitats such as the International Space Station.
Comparative genomics of bacteria associated with spacecraft cleanrooms: Genome analysis of Bacillus species producing radiation and peroxide resistant spores, which are biological contaminants in spacecraft assembly facilities and are also potential fouling hazards in nuclear facilities. This has relevance to how microbes evolve and adapt to the space environment if they find their way aboard unmanned and manned space missions.
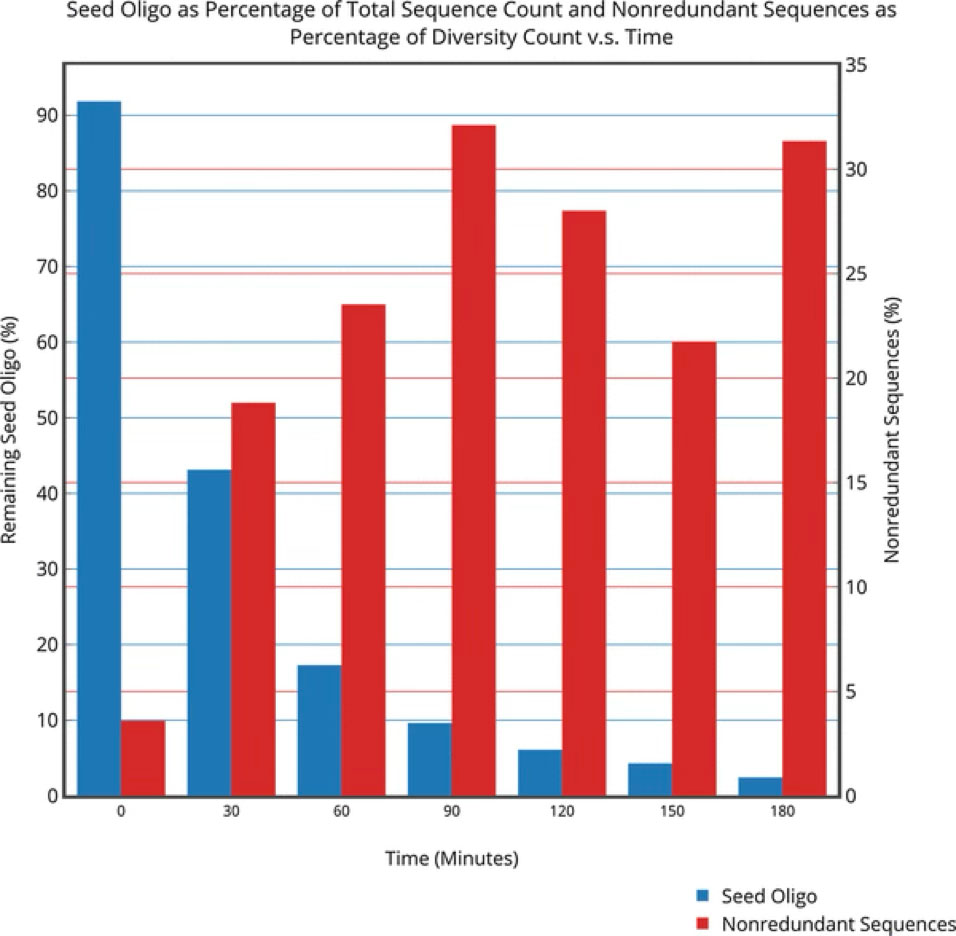
Understanding increase in RNA complexity and evolution, in the prebiotic milieu.
Understanding RNA structure, biology, and evolution, including the ribosome. The ribosome holds an important key to understand the emergence of coded protein synthesis in the context of the origins of life.
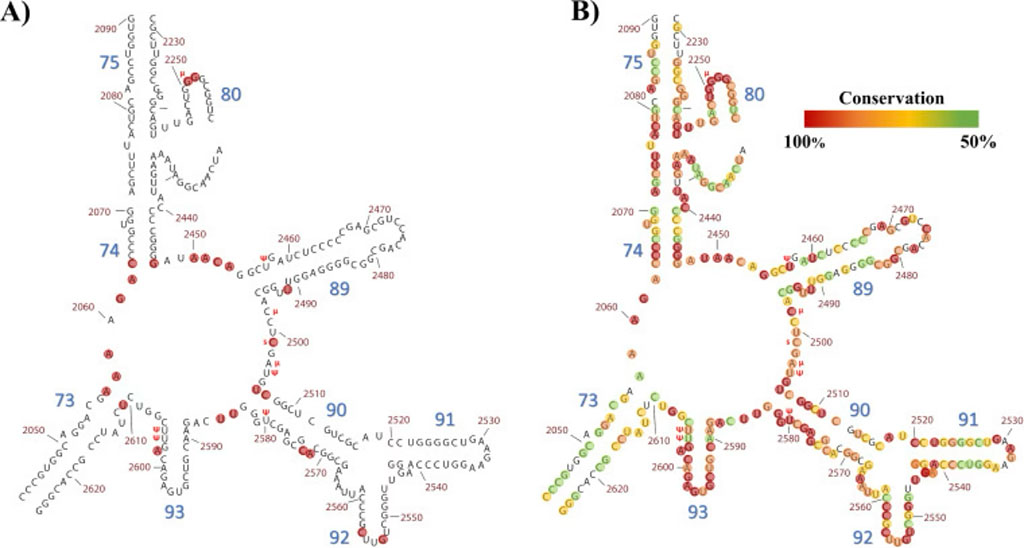
(B) Conservation of the nucleobases within this fragment according to crystallographic data and corresponding alignments from Bernier et al. (2014). This fragment comprises 282 nucleotides (nt). Red circles highlight those with 100% conservation (78 nt). Orange circles highlight those with 90 to 99.9% conservation (68 nt). Yellow and green circles highlight those with 70 to 89.9% and 50 to 69.9% conservation, respectively (52 and 49 nt). Thirty-five nucleotides from this fragment have shown less than 50% conservation.
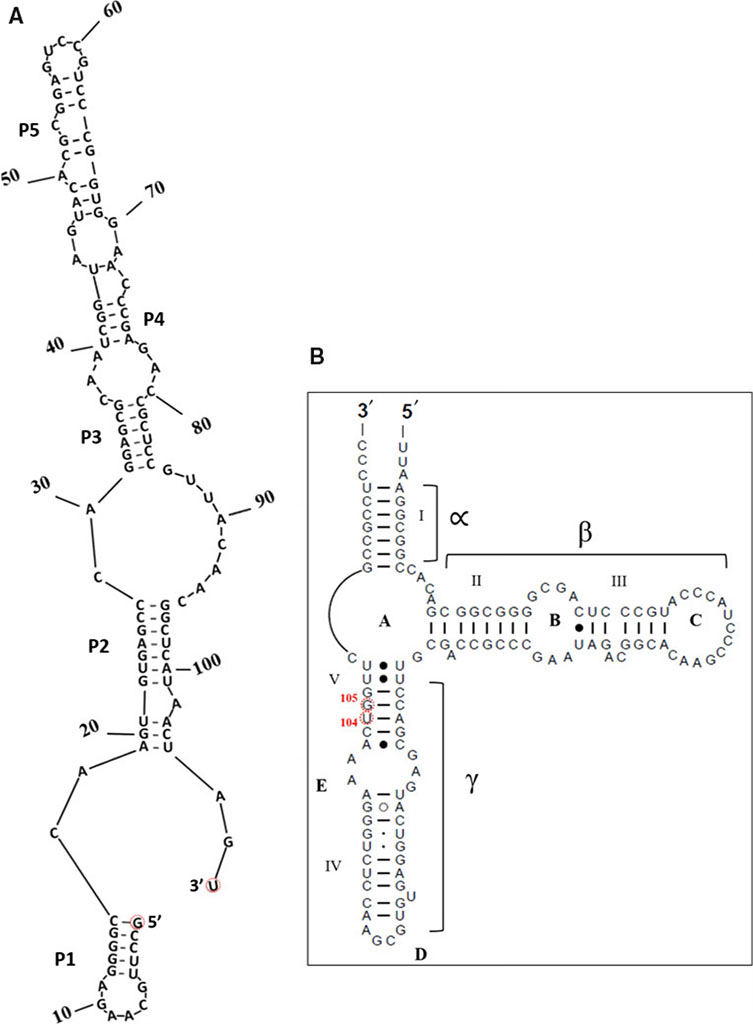
(A) a possible predicted secondary structure of the insert, and,
(B) the usual 5S rRNA secondary structure model (available at RNAcentral (https://rnacentral.org/) and the Comparative RNA Web (CRW) Site( http://www.rna.ccbb.utexas.edu)) and modified; The insert is between positions U104 and G105 as per Halococcus morrhuae 5S rRNA numbering. The equivalent positions in Haloarcula marismortui are C108 and G109. What would be the 5′ and 3′ ends of the insert if it were an independent RNA are indicated in (A).

(a) ΨThe orientation of all the genes in the entire cluster flipped for aligning with the other genomes. Each arrow represents a different location on a single complete scaffold; adjacent arrows do not indicate their order on the genome; genes within an arrow are contiguous. ¶Pseudogene; red diamond and red rectangle represent gene(s) absent in the corresponding genome/location; arrows within dashed red boxes are contiguous; HP: hypothetical protein; RNP1: ribonuclease P protein component 1; ORF: open reading frame/gene; #ORF(s) annotated as HP(s); Hsp20: heat shock protein 20; ßpartial gene. (b) ΨThe orientation of all the genes in the entire cluster flipped for aligning with the other genomes. In Thorarchaeota and Heimdallarchaeota, each colored (blue, green, or red) box represents a different contig; adjacent arrows do not indicate their order on the genome unless they are within a dashed red box; genes within an arrow are contiguous. ¶Pseudogene; red diamond represents gene(s) absent in the corresponding genome/location; arrows within dashed red boxes are contiguous; HP: hypothetical protein; RNP1: ribonuclease P protein component 1; ORF: open reading frame/gene; #ORF(s) annotated as HP(s); Hsp20: heat shock protein 20; ßpartial gene; IF Sui: protein translation factor SUI1 homolog; NCG: noncluster genes.
Journal Publications
- Tirumalai M. R.(2024) Education and public outreach: communicating science through storytelling. J Microbiol Biol Educ. 0:e00209-23. (https://journals.asm.org/doi/10.1128/jmbe.00209-23)
- Tirumalai M. R., Sivaraman R. V Jr, Kutty L. A., Song E. L., and Fox G. E. (2023), Ribosomal Protein Cluster Organization in Asgard Archaea. Archaea. Sep 29;2023:5512414. (https://doi.org/10.1155/2023/5512414). PMID: 38314098; PMCID: PMC10833476.
- Tirumalai M. R., Anane-Bediakoh D., Rajesh S., and Fox G. E. (2021), Net charges of the ribosomal proteins of the S10 and spc clusters of halophiles are inversely related to the degree of halotolerance. Microb Spectr. 9(3):e0178221, (https://doi.org/10.1128/spectrum.01782-21); PMID: 34908470; PMCID: PMC8672879.
- Tirumalai M. R., Rivas M., Tran Q., and Fox G. E. (2021), The Peptidyl Transferase Center: a Window to the Past. Microbiol Mol Biol Rev. Nov 10;85(4):e0010421, (https://doi.org/10.1128/MMBR.00104-21); PMID: 34756086; PMCID: PMC8579967.
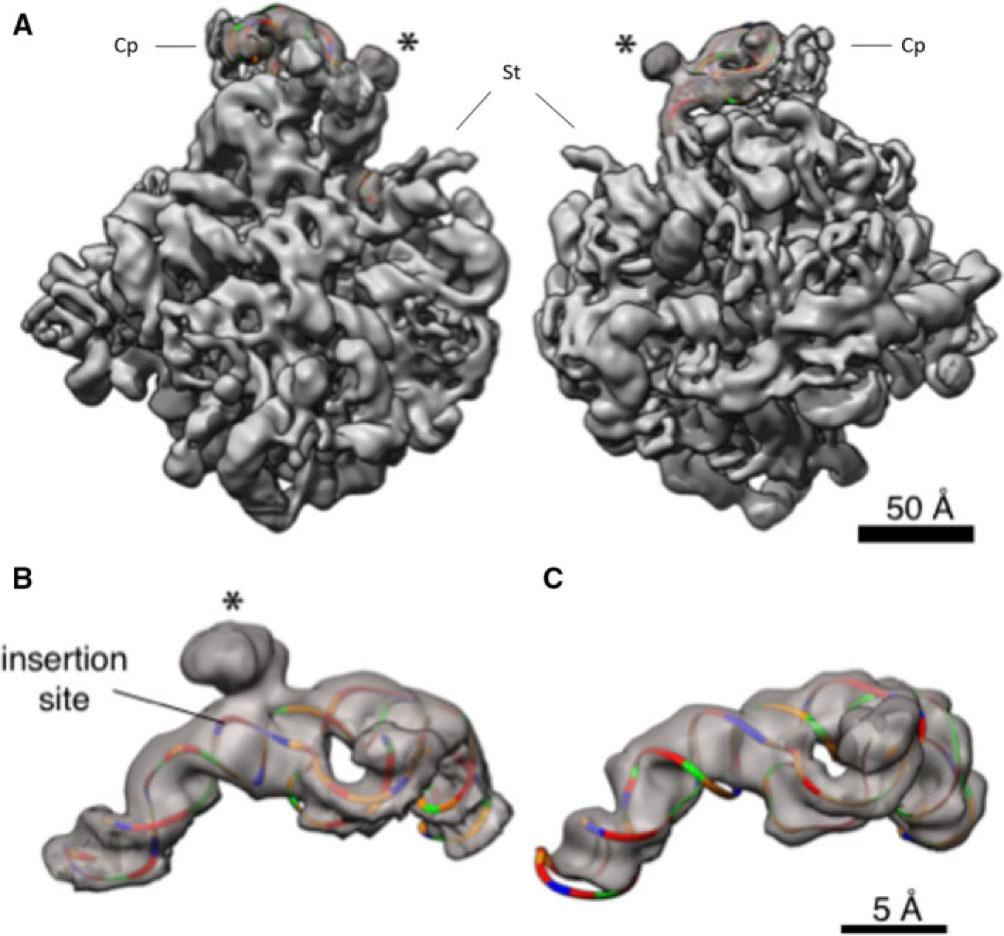
- Tirumalai M. R., Kaelber J. T., Park D. R., Tran Q., and Fox G. E. (2020), Cryo-electron microscopy visualization of a large insertion in the 5S ribosomal RNA of the extremely halophilic archaeon Halococcus morrhuae. FEBS Open Bio, 10: 1938-1946, (https://doi.org/10.1002/2211-5463.12962); PMID: 32865340, PMCID:PMC7530397.
- Camprubí E., de Leeuw J. W., House C. H., Raulin F., Russell M. J., Spang A., Tirumalai M. R., and Westall F. (2019), The Emergence of Life. Space Science Reviews, 215: 56, (https://doi.org/10.1007/s11214-019-0624-8). Contribution: Section “The RNA world – Reality or dogma?”
- Tirumalai M. R., Karouia F., Tran Q., Stepanov V. G., Bruce R. J., Ott C. M., Pierson D. L., and Fox G. E. (2019), Evaluation of Acquired Antibiotic Resistance in Escherichia coli Exposed to Long-Term Low-Shear Modeled Microgravity and Background Antibiotic Exposure. mBio, 10: e02637-18 (https://doi.org/10.1128/mBio.02637-18); PMID:30647159, PMCID:PMC6336426.
- Tirumalai M. R., Stepanov V. G., Wunsche A., Montazari S., Gonzalez R. O., Venkateswaran K., and Fox G. E. (2018), Bacillus safensis FO-36b and Bacillus pumilus SAFR-032: a whole genome comparison of two spacecraft assembly facility isolates. BMC Microbiology, 18: 57 (https://doi.org/10.1186/s12866-018-1191-y); PMID:29884123, PMCID: PMC5994023.
- Tirumalai M. R., Tran Q., Paci M., Chavan D., Marathe A., and Fox G. E. (2018), Exploration of RNA Sequence Space in the Absence of a Replicase. Journal of Molecular Evolution, 86: 264-276 (https://doi.org/10.1007/s00239-018-9846-8); PMID: 29748740.
- Tirumalai M. R., Karouia F., Tran Q., Stepanov V. G., Bruce R. J., Ott C. M., Pierson D. L., and Fox G. E. (2017), The adaptation of Escherichia coli cells grown in simulated microgravity for an extended period is both phenotypic and genomic. npj Microgravity, 3(15) (https://doi.org/10.1038/s41526-017-0020-1); PMID:28649637, PMCID: PMC5460176.
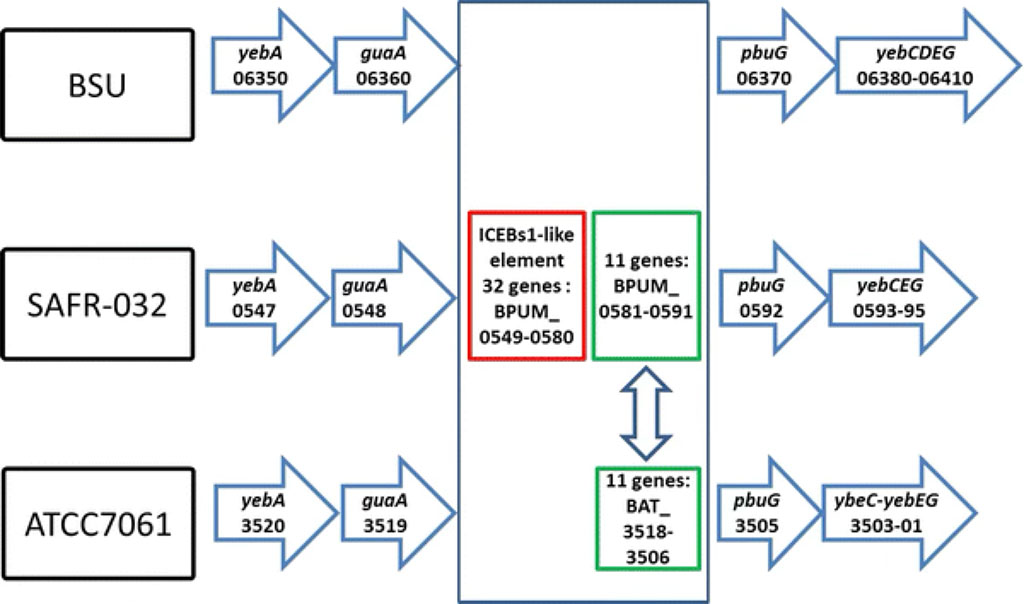
- Tirumalai M. R., Rastogi R., Zamani N., O'Bryant Williams E., Allen S., Diouf F., Kwende S., Weinstock G. M., Venkateswaran K. J., and Fox G. E. (2013), Candidate genes that may be responsible for the unusual resistances exhibited by Bacillus pumilus SAFR-032 spores. PLoS One, 8(6): e66012, (https://doi.org/10.1371/journal.pone.0066012); PMID: 23799069, PMCID: PMC3682946.
- Tirumalai M. R., and Fox G. E. (2013) An ICEBs1-like element may be associated with the extreme radiation and desiccation resistance of Bacillus pumilus SAFR-032 spores. Extremophiles, 17(5): 767-774 (https://doi.org/10.1007/s00792-013-0559-z); PMID: 23812891.
Book Contribution
- Camprubí E., de Leeuw J., House C., Raulin F., Russell M., Spang A., Tirumalai M. R., and Westall F. (2021) "Emergence of Life". In: Ocean Worlds: Habitability in the Outer Solar System and Beyond. edited by A Coustenis, Rodrigo, R., Spohn, T., Hand, K.P., Hayes, A., Olsson-Francis, K., Postberg, F., Sotin, C., Tobie, G., Raulin, F., Walter, N., L’Haridon, Js, Springer, Heidelberg, pp 29-82. Springer, Heidelberg, (Space Sciences Series of ISSI), Series Volume 77, Springer Netherlands (Hardcover ISBN 978-94-024-2069-2) (https://www.springer.com/us/book/9789402420692, http://www.issibern.ch/publications), Contribution: Section “The RNA world – Reality or dogma?”